BioJava:CookBook:PDB:alignGUI
A simple GUI for protein structure alignment
BioJava (in SVN) contains a simple GUI for easier working with protein structure alignments (<BioJava:CookBook:PDB:align>). The following code launches the user interface:
```java import org.biojava.bio.structure.gui.*;
public static void main(String[] args){
new AlignmentGui();
} ```
The AlignmentGui
In the user interface specify 2 PDB files (and optionally chain IDs) that should be superimposed.
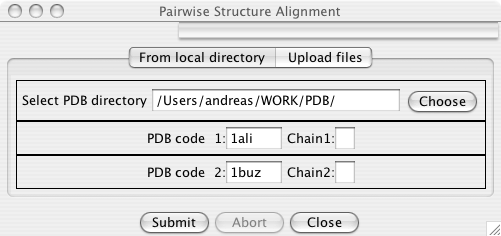
After pressing the Submit button, the alignment is calculated. A new frame pops up that shows the alternative solutions for this alignment:
Alternative Solutions
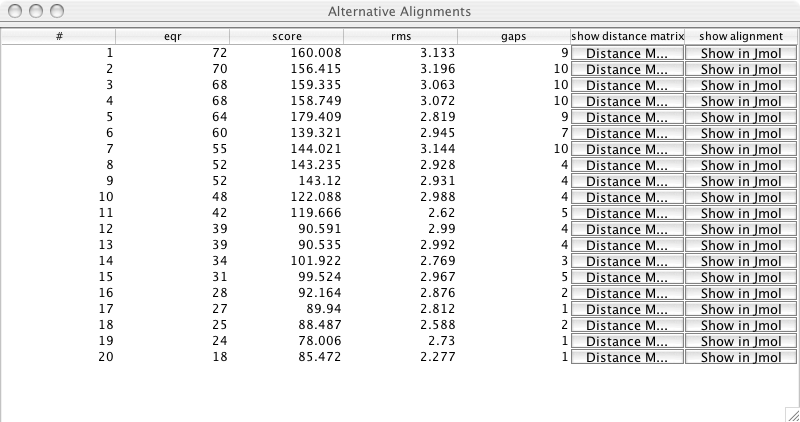
The columns in this table are:
#1 the number of the alternative alignment
eqr the number of structurally equivalent residues
score the score for this alternative alignment
rms root mean sqare
gaps number of gaps in the alignment
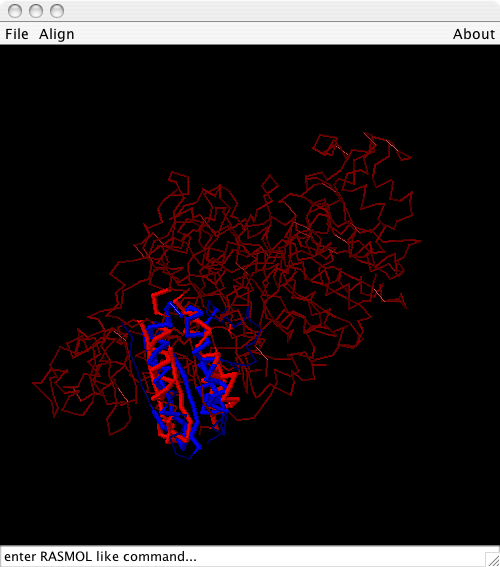
Display in Jmol
The Show in Jmol button allows to display this alternative alignment in Jmol, if it can be found on the classpath. If you don’t have Jmol installed, please get it from http://www.jmol.org. For more details on how to interact with Jmol see <BioJava:CookBook:PDB:Jmol>.
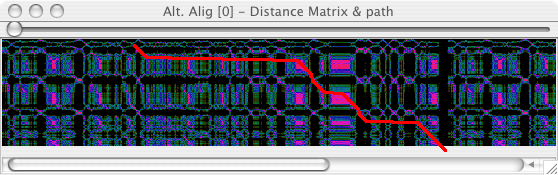
Internals of the algorithm
The Distance Matrix button shows the distance matrix that is used internally for the alignment and the path that has been choosen.
Configure PDB installation (Optional)
If you have a PDB installation that contains all PDB files in a single directory you can configure the System property PDB_DIR to point to this directory. (e.g. at startup specify -DPDB_DIR=path/to/PDB/files )