BioJava:CookBook:Interfaces:Alignments II
BioJava can render alignments
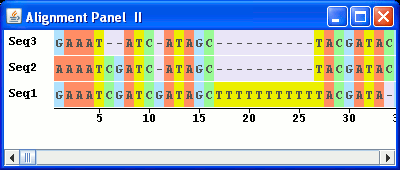
The AlignmentPanel_II example builds on the AlignmentPanel class. A scrollbar has been added to control the sequence on view. Also, LabelledSequenceRenderers have been useed to label each sequence in the the alignment. Lastly, the look of the sequences has been modifed.
To allow this the paint method of the SymbolSequenceRenderer class has been overridden to allow control of the sequence being displayed. Each nucleotide is framed in a rectangle filled with colour according to the base.
The following is a screenshot of the viewer generated by the AlignmentPanel_II class:
```java //Load the Java libraries import java.awt.*; import java.awt.event.*; import java.awt.geom.*; import java.util.*; import javax.swing.*; //Load the BioJava libraries import org.biojava.bio.*; import org.biojava.bio.seq.*; import org.biojava.bio.seq.io.*; import org.biojava.bio.symbol.*; import org.biojava.bio.gui.sequence.*;
public class AlignmentPanel_II extends JFrame {
//Create references to the sequences
Sequence seq, seq1, seq2, seq3;
//Instantiate the BioJava GUI elements
//TranslatedSequencePanel to hold the renderers
TranslatedSequencePanel tsp = new TranslatedSequencePanel();
//LabelledSequenceRenderer for each AlignmentRenderer
LabelledSequenceRenderer labRen1, labRen2, labRen3;
//AlignmentRenderer to hold each sequence
AlignmentRenderer render1, render2, render3;
//MultiLineRenderer to allow display of multiple tracks in the TranslatedSequencePanel
MultiLineRenderer multi = new MultiLineRenderer();
//SymbolSequenceRenderer to handle display of the sequence symbols - only one instance is needed
SymbolSequenceRenderer symbol = new SymbolSequenceRenderer();
//RulerRenderer to display sequence coordinates
RulerRenderer ruler = new RulerRenderer();
//The width in pixels of the of the label in the LabelledSequenceRenderer
int labelWidth = 50;
//The height in pixels of the of the label in the LabelledSequenceRenderer
int labelHeight = 25;
JScrollBar scrollBar;
public AlignmentPanel_II(){
super("Alignment Panel II");
//Create the sequences for the alignment
try {
seq1 = DNATools.createGappedDNASequence("GAAATCGATCGATAGCTTTTTTTTTTTACGATA-GACTAGCATTCCGACGATA-GACTAGCATTCCC", "Seq1");
seq2 = DNATools.createGappedDNASequence("AAAATCGATC-ATAGC----------TACGATACGACTAGCATTCCGAC--TA-GACTAGCATTCC-", "Seq2");
seq3 = DNATools.createGappedDNASequence("GAAAT--ATC-ATAGC----------TACGATACGACTAGCATTCCGAC--TA--ACTAGG----CC", "Seq3");
}
catch (BioException bioe) {
System.err.println("Bioexception: " + bioe);
}
//Overide the paint method of the SymbolSequenceRenderer class to allow modification of the sequence being displayed
//To do this you will also need to modify the access level of the double depth and the Paint outline variables.
//They are private so either change that or add a get method for each.
SymbolSequenceRenderer symbol = new SymbolSequenceRenderer(){
public void paint(Graphics2D g2, SequenceRenderContext context) {
Rectangle2D prevClip = g2.getClipBounds();
AffineTransform prevTransform = g2.getTransform();
g2.setPaint(outline);
Font font = context.getFont();
Rectangle2D maxCharBounds = font.getMaxCharBounds(g2.getFontRenderContext());
double scale = context.getScale();
if (scale >= (maxCharBounds.getWidth() * 0.3) && scale >= (maxCharBounds.getHeight() * 0.3)) {
double xFontOffset = 0.0;
double yFontOffset = 0.0;
xFontOffset = maxCharBounds.getCenterX() * 0.25;
yFontOffset = - maxCharBounds.getCenterY() + (depth * 0.5);
SymbolList seq = context.getSymbols();
SymbolTokenization toke = null;
try {
toke = seq.getAlphabet().getTokenization("token");
}
catch (Exception ex) {
throw new BioRuntimeException(ex);
}
Location visible = GUITools.getVisibleRange(context, g2);
for (int sPos = visible.getMin(); sPos <= visible.getMax(); sPos++) {
double gPos = context.sequenceToGraphics(sPos);
String s = "?";
try {
s = toke.tokenizeSymbol(seq.symbolAt(sPos));
}
catch (Exception ex) {
// We'll ignore the case of not being able to tokenize it
}
//Start of the modifications -------------------------------
//Make sure the text is uppercase
s = s.toUpperCase();
//Set the color according to the nucleotide for the background
if (s.equals("A")){g2.setColor(new Color(255,140,105));}
else if (s.equals("T")){g2.setColor(new Color(238,238,0));}
else if (s.equals("G")){g2.setColor(new Color(176,226,255));}
else if (s.equals("C")){g2.setColor(new Color(151,251,152));}
else {g2.setColor(new Color(230,230,250));}
//Plot the rectangle to frame the nucleotide symbol
g2.fill(new Rectangle2D.Double((gPos + xFontOffset)-1.5, 0, tsp.getScale(), labelHeight ));
//Set the colour for the text
g2.setColor(new Color(83,83,83));
//End of the modifications ---------------------------------
g2.drawString(s, (float)(gPos + xFontOffset), (float)yFontOffset);
}
}
g2.setTransform(prevTransform);
}
};
//Use the Map to create a new SimpleAlignment
Map<String, Sequence> list = new HashMap();
list.put(seq1.getName(), seq1);
list.put(seq2.getName(), seq2);
list.put(seq3.getName(), seq3);
SimpleAlignment ali = new SimpleAlignment((Map) list);
//Instantiate the AlignmentRenderer
render1 = new AlignmentRenderer();
//Set the label for the AlignmentRenderer
render1.setLabel(ali.getLabels().get(0));
//Set the renderer for the AlignmentRenderer
render1.setRenderer(symbol);
//Instantiate the LabelledSequenceRenderer
labRen1 = new LabelledSequenceRenderer(labelWidth, labelHeight);
//Set the name of the sequence as the label in the LabelledSequenceRenderer
labRen1.addLabelString(render1.getLabel().toString());
//Put the AlignmentRenderer in the LabelledSequenceRenderer
labRen1.setRenderer(render1);
render2 = new AlignmentRenderer();
render2.setLabel(ali.getLabels().get(1));
render2.setRenderer(symbol);
labRen2 = new LabelledSequenceRenderer(labelWidth, labelHeight);
labRen2.addLabelString(render2.getLabel().toString());
labRen2.setRenderer(render2);
render3 = new AlignmentRenderer();
render3.setLabel(ali.getLabels().get(2));
render3.setRenderer(symbol);
labRen3 = new LabelledSequenceRenderer(labelWidth, labelHeight);
labRen3.addLabelString(render3.getLabel().toString());
labRen3.setRenderer(render3);
//Add the alignment renderers to the MultiLineRenderer
multi.addRenderer(labRen1);
multi.addRenderer(labRen2);
multi.addRenderer(labRen3);
//Add the ruler to the MultiLineRenderer
multi.addRenderer(ruler);
//Set the sequence in the TranslatedSequencePanel
tsp.setSequence((SymbolList)ali);
//Set the background colour of the TranslatedSequencePanel
tsp.setOpaque(true);
tsp.setBackground(Color.white);
//Set the renderer for the TranslatedSequencePanel
tsp.setRenderer(multi);
//Create a scrollbar and add an adjustment listener
scrollBar = new JScrollBar(JScrollBar.HORIZONTAL, 0, 0, 0, 100);
scrollBar.addAdjustmentListener(
new AdjustmentListener() {
public void adjustmentValueChanged(AdjustmentEvent e) {
//Get the absolute position of the scroll bar
double scrollBarValue = e.getValue();
//Get the position of the scroll bar relative to the maximum value
double scrollBarRatio = scrollBarValue / scrollBar.getMaximum();
//Calculate the new position of the first base to be displayed
double pos = scrollBarRatio * (tsp.getSequence().length() - ((getWidth() - labelWidth) / tsp.getScale()));
//Set the new SymbolTranslation for the TranslatedSequencePanel
tsp.setSymbolTranslation((int)Math.round(pos));
}
}
);
//Set up the display
Container con = getContentPane();
con.setLayout(new BorderLayout());
con.add(tsp, BorderLayout.CENTER);
con.add(scrollBar, BorderLayout.SOUTH);
setSize(400,170);
setLocation(100,100);
setVisible(true);
setDefaultCloseOperation(WindowConstants.EXIT_ON_CLOSE);
}
/**
* Main method
*/
public static void main(String args []){
new AlignmentPanel_II();
}
} ```