BioJava:Tutorial:Symbols and SymbolLists
Tutorial
By Thomas Down
This chapter covers the fundamentals of accessing biological sequence
data from BioJava, and explains how BioJava’s treatment of sequences
differs from other libraries. This chapter refers to Java API defined in
the packages org.biojava.bio.symbol and org.biojava.bio.seq. For a
complete overview of the APIs provided by these packages, please consult
the JavaDoc API documentation (latest biojava
1.8).
Symbols and Alphabets
When biological sequence data first became available, it was necessary to find a convenient way to communicate it. A logical approach is to represent each monomer in a biological macromolecule using a single letter - usually the initial letter of the chemical entity being described, for instance ‘T’ for thymidine residues in DNA. When this data was entered into computers, it was logical to use the same scheme. A lot of computational biology software is based on normal string handling APIs. While the notion of a sequence as a string of ASCII characters has served us well to date, there are several issues which can present problems to the programmer:
Validation: It is possible to pass any string to a routine which is expecting a biological sequence. Any validation has to be performed on an ad hoc basis.
Ambiguity: The meaning of each symbol is not necessarily clear. The ‘T’ which means thymidine in DNA is the same ‘T’ which is a threonine residue in a protein sequence
Limited alphabet: While there are obvious encodings for nucleic acid and sequence data as strings, the same approach does not always work well for other kinds of data generated in biological sequence analysis software
BioJava takes a rather different approach to sequence data. Instead of
using a string of ASCII characters, a sequence is modelled as a list of
Java objects implementing the Symbol interface. This class, and the
others described here, are part of the Java package
org.biojava.bio.symbol.
public interface Symbol {
public String getName();
public Annotation getAnnotation();
public Alphabet getMatches();
}
All Symbol instances have a name property (for instance, Thymidine).
They may optionally have extra information associated with them (for
instance, information about the chemical properties of a DNA base)
stored in a standard BioJava data structure called an Annotation.
Annotations are just set of key-value data. The final method,
getMatches, is only important for ambiguous symbols, which are covered
at the end of this chapter.
The set of Symbol objects which may be found in a particular type of
sequence data are defined in an Alphabet. It is always possible to
define custom symbols and alphabets, but BioJava supplies a set of
predefined alphabets for representing biological molecules. These are
accessible through a central registry called the AlphabetManager, and
through convenience methods.
FiniteAlphabet dna = DNATools.getDNA();
Iterator dnaSymbols = dna.iterator();
while (dnaSymbols.hasNext()) {
Symbol s = (Symbol) dnaSymbols.next();
System.out.println(s.getName());
}
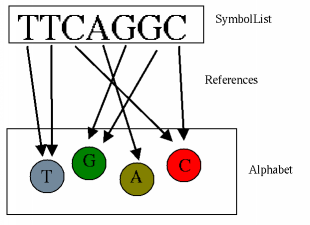
SymbolList: the simple sequence
The basic interface for sequence data in BioJava is SymbolList. Every
symbol list has an associated alphabet, and may only contain symbols
from that alphabet. Symbol lists can be seen as strings which are made
up of Symbol objects rather than characters. The interface specifies
methods for querying the alphabet and length, and accessing the symbols:
SymbolList seq = getSomeSequence();
System.out.println("Alphabet = " + seq.getAlphabet().getName());
System.out.println("Length = " + seq.length());
System.out.println("First symbol = " + seq.symbolAt(1).getName());
Note that numbering of symbols within the symbol list runs from 1 to
length, not from 0 to length - 1 as is the case with Java strings.
This is consistent with the coordinate system found in files of
annotated biological sequences.
There are several other standard methods in the SymbolList interface.
subList returns a new symbol list representing part of the sequence,
just like the substring method of the String class. seqString
returns a normal string representation of the sequence. This latter
method will only work if the symbol list uses an alphabet where all
symbols have their token property defined. However, since this is true
of the commonly used DNA and protein alphabets, this method is useful if
you need interaction between BioJava and legacy sequence analysis code.
The SymbolList interface does not define any methods for modifying the
underlying sequence data. Future versions of BioJava may also include a
MutableSymbolList interface.
Doesn’t this all waste memory?
A common concern with BioJava’s Symbol/SymbolList model is that it
must use much more memory than a simple string-based approach to
sequence storage. It should be stressed that BioJava does not use a
separate object to represent each nucleotide in a long DNA sequence. In
fact, there are just four ‘singleton’ Symbol objects which represent
the symbols found in the DNA alphabet. These can be accessed at any time
using static methods of the DNATools class. Whenever a thymidine
residue is stored in a sequence, all that is really stored is a
reference to the singleton thymidine object. Typically, this takes up
four bytes of memory: more than the two bytes used by a Java char, but
still manageable.
Actually, it is possible in principle to store a DNA sequence (without
gaps or ambiguous residues) using only two bits per residue. Since the
BioJava SymbolList is an interface, it only defines how the sequence
should be accessed - not how data is stored. If space is important, it
is possible to implement a ‘packed’ implementation of SymbolList.
Client code need never worry about the underlying data model.
BioJava’s object oriented view of sequences brings other advantages. Many programs which analyse DNA sequences need to have simultaneous access to the original sequence and that of its complementary strand. In BioJava this is easy.
SymbolList forward = getSequence();
SymbolList backward = DNATools.reverseComplement(forward);
System.out.println("First base: " + forward.symbolAt(1).getName());
System.out.println("Complement: " + backward.symbolAt(backward.length()).getName());
Since the reverse complement of a DNA sequence is a simple programmatic
transformation, BioJava doesn’t need to physically store the sequence in
memory at all. Instead, it just creates a special implementation of the
SymbolList interface, which computes the reverse strand sequence on
the fly. This will typically cost just a few bytes of memory regardless
of the sequence length, compared to megabytes for a string
representation of a typical genome sequence.
How do I access my sequence data?
Each Alphabet object can have one or more SymbolTokenization
implementations associated. These are two-way mappings between Symbol
objects and textual representations of the data. They are the primary
mechanism for creating new symbol lists from existing
(character-encoded) sequence data. By convention, any alphabet which has
a commonly accepted textual representation has a symbol tokenization
called ‘token’ associated:
String seqString = "GATTACA";
Alphabet dna = DNATools.getDNA();
SymbolTokenization dnaToke = dna.getTokenization("token");
SymbolList seq = new SimpleSymbolList(dnaToke, seqString);
String seqString2 = dnaToke.tokenizeSymbolList(seq);
System.out.println("Strings match: " + seqString2.equalsIgnoreCase(seqString));
This low-level parsing mechanism is supplemented by a more sophisticated
sequence Input/Output framework, defined in the package
org.biojava.bio.seq.io. This uses pluggable file format converters,
and can currently read and write in Fasta, EMBL, and Genbank formats.
BioJava can also fetch data from services such as DAS using
Dazzle, and access databases
such as Genbank and BioSQL as well those used by the
Ensembl project (additional packages are
required to support DAS and Ensembl).
What about the Sequence interface?
Until this point, we have concentrated on the SymbolList interface
which, as its name suggests, is a raw list of Symbol references. Real
entries in sequence databases are more complicated than this: sequences
almost always have some kind of ID code or description associated, and
many are also accompanied by tables of annotations. In BioJava,
Sequence is a subinterface of SymbolList which adds a name
property, plus a mechanism for querying tables of features.
The general rule is that the Sequence interface is normally used for
sequences which have been loaded into a program from files or databases.
SymbolList may be a more appropriate type for sequences generated
internally by an analysis program.
A simple example
The following program is a very simple example, which reads one or more DNA sequences from a FASTA format data file and reports the GC content of each. This example is a (very) simple application of the BioJava Sequence I/O framework, described in later chapters. Used as below, it allows you to iterate over all the sequences in a multiple-entry file, rather than holding all of them in memory at once.
```javaimport java.io.*; import org.biojava.bio.symbol.*; import org.biojava.bio.seq.*; import org.biojava.bio.seq.io.*;
public class GCContent {
public static void main(String[] args)
throws Exception
{
if (args.length != 1)
throw new Exception("usage: java GCContent filename.fa");
String fileName = args[0];
// Set up sequence iterator
BufferedReader br = new BufferedReader(
new FileReader(fileName));
SequenceIterator stream = SeqIOTools.readFastaDNA(br);
// Iterate over all sequences in the stream
while (stream.hasNext()) {
Sequence seq = stream.nextSequence();
int gc = 0;
for (int pos = 1; pos <= seq.length(); ++pos) {
Symbol sym = seq.symbolAt(pos);
if (sym == DNATools.g() || sym == DNATools.c())
++gc;
}
System.out.println(seq.getName() + ": " +
((gc * 100.0) / seq.length()) +
"%");
}
}
}```
Ambiguous symbols
Sometimes, it is useful to represent sequences which are not perfectly defined. In such cases, it is common to use ambiguous symbols. A common example is the ‘N’ character in DNA sequences, which is used to indicate parts of a sequence where the sequencing traces were difficult to interpret. Sometimes, runs of Ns are also used to indicate gaps in assemblies. In the case of DNA, additional ambiguity symbols have been defined, covering all possible combinations of the four bases. For instance, the symbol ‘W’ realy means (A or T).
Within the BioJava object model, it is possible to inspect any ambiguous
symbol to determine the set of atomic symbols which it matches, using
the getMatches method. Atomic symbols can be considered to be the
special case where getMatches returns a set whose size is exactly one.
As a conveniece, atomic symbols also implement the AtomicSymbol
interfaces.
You might want to modify the GCContent program, above, so as to ignore any ambiguous symbols in the input sequence.