BioJava:CookBook:Interfaces:ProteinPeptideFeatures
How do I display protein features / a Peptide Digest?
Note: this recipe makes use of the latest BioJava version available via CVS.
This example demonstrates several different features of the ProteinDigestDemo. To best view this demo you will need a SwissProt format file that contains secondary structural elements in its Feature Table. The following link is for such a file:
http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-id+465_c1S9c9A+-e+[SWISSPROT:'PPARG_HUMAN']+-qnum+1+-enum+2
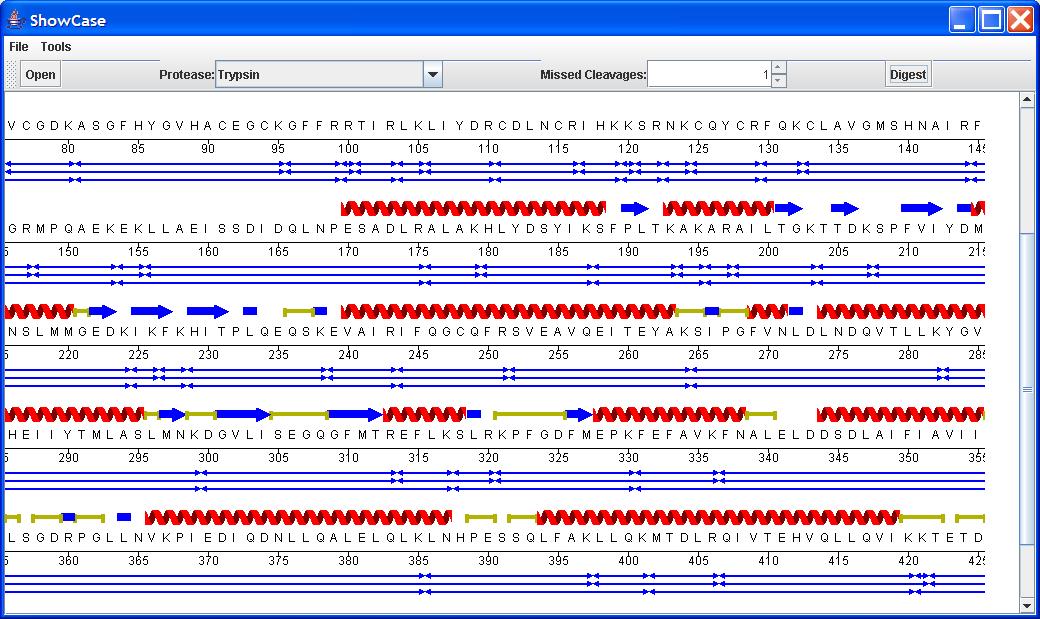
1) Wrapped Sequence Display
Proteins are typically viewed at or close to single residue resolution so we use an org.biojava.bio.gui.sequence.SequencePanelWrapper that renders the sequence over several horizontal or vertical tracks in the display. In this way, at residue resolution we can see far more of the sequence on screen and it looks far more like something we could print or publish.
The SequencePanelWrapper makes use of different layout strategies via classes that implement the org.biojava.bio.gui.sequence.tracklayout.TrackLayout interface. A simple TrackLayout strategy is to render the same number of residues per line. A more complicated strategy is to render different numbers of residues on each line. This might be useful in cases where you would not want to break up the rendering of a feature across lines.
2) An offset ruler
For when a sequence starts at a position other than +1. The example is when your protein contains a his tag that would otherwise make the sequence coordinates incompatible.
3) Display of Secondary Structural Features (Helices, Turns, Sheets) and Domains
A Swissprot sequence file when loaded may include secondary structural features from it's Feature Table. Here we make use of org.biojava.bio.gui.sequence.GlyphFeatureRenderer's and subclasses such as SecondaryStructureFeatureRenderer that render glyphs (HelixGlyph, TurnGlyph etc.) from the org.biojava.bio.gui.glyph package and are implementors of the org.biojava.bio.gui.glyph.Glyph interface.
4) Display of a Protein Digest
This example brings together the sequence gui and the org.biojava.bio.proteomics package. The key class is an org.biojava.bio.gui.sequence.PeptideDigestRenderer. We use the Digest class from the proteomics package to generate sequence features of the Digest.PEPTIDE_FEATURE_TYPE and then filter for these with the PeptideDigestRenderer. The PeptideDigestRenderer is a subclass of MultiLineRenderer and internally sorts and aligns the features so that they do not overlap in the display, creating extra lines as necessary. The rendering of the features is very configurable with the parent class method: public
FeatureRenderer createRenderer(int
lane) overridden for custom rendering.
```java import org.biojava.bio.*; import org.biojava.bio.symbol.*; import org.biojava.bio.gui.sequence.*; import org.biojava.bio.gui.sequence.tracklayout.*; import org.biojava.bio.gui.glyph.*; import org.biojava.bio.seq.*; import org.biojava.bio.seq.impl.*; import org.biojava.bio.seq.io.*; import org.biojava.utils.*; import org.biojava.bio.proteomics.*;
import java.io.*; import javax.swing.*; import java.awt.*; import java.awt.event.*; import java.util.*;
/**
*
PeptideDigestDemo demonstrates the use of several new
SequenceRenderers and a couple of sequence layouts.
* The SequencePanelWrapper class allows a page like view of a
* If a Swissprot format sequence file with structural features is imported then
* Alpha Helices, Beta Sheets and Domains will be rendered.
* The functionality of the org.biojava.bio.proteomics package is also demonstrated via the
* PeptideDigestRenderer.
*
*
* @author Mark Southern
* @since 1.4
*/
public class PeptideDigestDemo extends JFrame{
private MultiLineRenderer multi;
private SequencePanelWrapper sequencePanel;
private Sequence seq;
private PeptideDigestRenderer digestRenderer;
private OffsetRulerRenderer offsetRenderer;
private JToolBar toolBar;
private JMenuBar menuBar;
public PeptideDigestDemo(){
setTitle("ShowCase");
configureSequencePanel();
Action action = new OpenSequenceAction();
toolBar = new JToolBar();
getContentPane().add(toolBar, BorderLayout.NORTH);
toolBar.add( new JButton( action ) );
toolBar.add( new JSeparator());
menuBar = new JMenuBar();
setJMenuBar(menuBar);
JMenu menu = new JMenu("File");
menuBar.add(menu);
menu.add( new JMenuItem( action ) );
configureProteaseCombo();
menu = new JMenu("Tools");
menuBar.add(menu);
action =new OffsetAction();
menu.add(new JMenuItem( action ));
action =new SmoothTrackWrapAction();
menu.add(new JMenuItem( action ));
action = new UserDefinedTrackWrapAction();
menu.add(new JMenuItem( action ));
setDefaultCloseOperation(WindowConstants.DISPOSE_ON_CLOSE);
getContentPane().add(new JScrollPane(sequencePanel), java.awt.BorderLayout.CENTER);
pack();
setSize(800, 800);
}
protected void configureSequencePanel(){
sequencePanel = new SequencePanelWrapper();
sequencePanel.setSequence(seq);
MultiLineRenderer multi = new MultiLineRenderer();
sequencePanel.setRenderer(multi);
try{
multi.addRenderer( createDomainRenderer() );
multi.addRenderer( createSecondaryStructureRenderer() );
multi.addRenderer(new SymbolSequenceRenderer());
multi.addRenderer( offsetRenderer = new OffsetRulerRenderer());
multi.addRenderer( createPeptideDigestRenderer() );
}
catch(ChangeVetoException ex){
ex.printStackTrace();
}
}
protected void configureProteaseCombo(){
final JComboBox proteaseCombo = new JComboBox( new DefaultComboBoxModel() );
Object selected = proteaseCombo.getSelectedItem();
((DefaultComboBoxModel)proteaseCombo.getModel()).removeAllElements();
int idx = -1;
int i = 0;
for(Iterator it = new TreeSet( ProteaseManager.getNames() ).iterator(); it.hasNext(); ){
String protease = (String)it.next();
if( protease.equals(selected))
idx = i;
i++;
proteaseCombo.addItem(protease);
}
toolBar.add( new JLabel("Protease:") );
toolBar.add( proteaseCombo );
toolBar.add( new JSeparator());
toolBar.add( new JLabel("Missed Cleavages:"));
final JSpinner missedCleavages = new JSpinner( new SpinnerNumberModel(0,0,10,1));
toolBar.add( missedCleavages );
toolBar.add( new JSeparator());
JButton b = new JButton( new AbstractAction("Digest"){
public void actionPerformed(ActionEvent e){
try{
ViewSequence view = new ViewSequence(seq);
Digest digest = new Digest();
digest.setSequence( view );
String proteaseName = proteaseCombo.getSelectedItem().toString();
digest.setProtease( ProteaseManager.getProteaseByName(proteaseName) );
int max = ((Integer)missedCleavages.getValue()).intValue();
digest.setMaxMissedCleavages(max);
digest.addDigestFeatures();
setViewSequence(view);
digestRenderer.sortPeptidesIntoLanes();
}
catch(Exception ex){
JOptionPane.showMessageDialog((Component)e.getSource(),"There was an error digesting the protein","Demo", JOptionPane.ERROR_MESSAGE);
}
}
});
toolBar.add(b);
toolBar.add( new JSeparator());
}
protected SequenceRenderer createSecondaryStructureRenderer() throws ChangeVetoException{
SecondaryStructureFeatureRenderer fr = new SecondaryStructureFeatureRenderer();
FeatureBlockSequenceRenderer block = new FeatureBlockSequenceRenderer();
block.setFeatureRenderer(fr);
return block;
}
protected SequenceRenderer createDomainRenderer() throws ChangeVetoException{
GlyphFeatureRenderer gfr = new GlyphFeatureRenderer();
gfr.addFilterAndGlyph(new FeatureFilter.ByType("DOMAIN"),
new TurnGlyph(java.awt.Color.GREEN.darker(), new java.awt.BasicStroke(3F))
);
FeatureBlockSequenceRenderer block = new FeatureBlockSequenceRenderer();
block.setFeatureRenderer(gfr);
return block;
}
protected SequenceRenderer createPeptideDigestRenderer() throws ChangeVetoException{
digestRenderer = new PeptideDigestRenderer( new FeatureSource(){
public FeatureHolder getFeatureHolder(){
return sequencePanel.getSequence();
}
});
digestRenderer.setFilter( new FeatureFilter.ByType( Digest.PEPTIDE_FEATURE_TYPE ) );
return digestRenderer;
}
protected void setViewSequence(ViewSequence seq){
sequencePanel.setSequence(seq);
}
public static void main(String[] args) throws IOException, BioException, ChangeVetoException{
PeptideDigestDemo s = new PeptideDigestDemo();
s.setVisible(true);
}
class OpenSequenceAction extends AbstractAction{
public OpenSequenceAction(){
super("Open");
}
public void actionPerformed(ActionEvent e){
JFileChooser chooser = new JFileChooser();
int result = chooser.showOpenDialog((Component)e.getSource());
if( result != JFileChooser.APPROVE_OPTION )
return;
File f = chooser.getSelectedFile();
try{
SequenceIterator iter = ( SequenceIterator ) SeqIOTools.fileToBiojava(SeqIOTools.guessFileType(
f), new BufferedReader(new FileReader(f))
);
seq = iter.nextSequence();
setViewSequence(new ViewSequence(seq));
}
catch(Exception ex){
JOptionPane.showMessageDialog((Component)e.getSource(), "There was an error opening the sequence","Demo", JOptionPane.ERROR_MESSAGE);
}
}
}
class OffsetAction extends AbstractAction{
public OffsetAction(){
super("Set Ruler Offset");
}
public void actionPerformed(ActionEvent e){
String result = JOptionPane.showInputDialog((Component)e.getSource(), "Enter an offset for the ruler","Demo", JOptionPane.QUESTION_MESSAGE);
try{
int i = Integer.parseInt(result);
offsetRenderer.setSequenceOffset(i);
}
catch(Exception ex){
JOptionPane.showMessageDialog((Component)e.getSource(), "There was an error setting the ruler","Demo", JOptionPane.ERROR_MESSAGE);
}
}
}
class SmoothTrackWrapAction extends AbstractAction{
public SmoothTrackWrapAction(){
super("Smooth Track Wrapping");
}
public void actionPerformed(ActionEvent e) {
String result = JOptionPane.showInputDialog((Component)e.getSource(),
"Enter a single value on which to wrap");
try{
int i = Integer.parseInt(result);
sequencePanel.setTrackLayout(new SimpleTrackLayout(sequencePanel.getSequence(),i));
}
catch(Exception ex){
JOptionPane.showMessageDialog((Component)e.getSource(), "There was an error setting the wrapping","Demo", JOptionPane.ERROR_MESSAGE);
}
}
}
class UserDefinedTrackWrapAction extends AbstractAction{
public UserDefinedTrackWrapAction(){
super("Set User Defined Track Wrapping");
}
public void actionPerformed(ActionEvent e) {
TrackLayout tl = sequencePanel.getTrackLayout();
RangeLocation[] ranges = tl.getRanges();
String expr = "";
for (int i = 0; i < ranges.length; i++) {
expr += ranges[i].getMax();
if (i < ranges.length) {
expr += ",";
}
}
expr = JOptionPane.showInputDialog((Component)e.getSource(),
"Enter the values on which to wrap (comma separated)", expr
);
if (expr == null) {
return;
}
String[] nums = expr.split("[\\s,\\t]+");
ranges = new RangeLocation[nums.length];
int min = 1;
for (int i = 0; i < nums.length; i++) {
int max = Integer.parseInt(nums[i]);
ranges[i] = new RangeLocation(min, max);
min = max + 1;
}
sequencePanel.setTrackLayout(new UserDefinedTrackLayout(ranges));
}
}
} ```