BioJava:CookBook:PDB:bioassembly
Since BioJava 3.0.5 the protein structure modules can parse the biological assembly information out of PDB and mmcif files and create a biologically correct representation of protein structures.
For more information about the differences between the asymmetric unit and the biological assembly see a tutorial at RCSB PDB
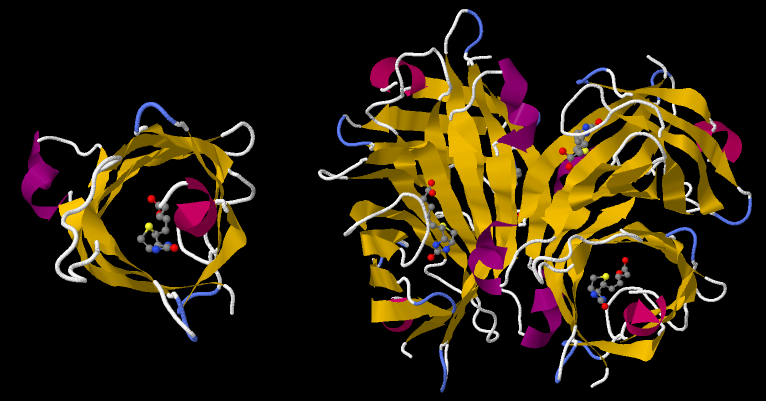
An comparison of the asymmetric unit and the biological assembly of Streptavidin is shown in this screenshot: It compares the asymmetric unit of PDB ID 1STP (left side) with its biological assembly (right side).
Here some example code how to load the biological assembly:
// good examples: 1stp 1gav 1hv4 1hho 7dfr 3fad 1qqp
// assembly 0 ... asym Unit
// assembly 1 ... the first bio assembly
// example 1fah has 2 assemblies (two copies of the assembly in asymmetric unit)
// Various interesting symmetries: (see Lawson, 2008)
// Circular - 1TJA
// Dihedral - 1ei7
// Icosahedral - 1a34
// Helical - 1cgm
// DNA 173D .. 2
// we load the bio assembly info out of the PDB files
BioUnitDataProviderFactory.setBioUnitDataProvider(BioUnitDataProviderFactory.pdbProviderClassName);
//Structure bioAssembly = StructureIO.getBiologicalAssembly("4A1I",2);
Structure bioAssembly = StructureIO.getBiologicalAssembly("1ei7",1);
StructureAlignmentJmol jmolPanel = new StructureAlignmentJmol();
//jmolPanel.evalString("set autobond=false");
jmolPanel.setStructure(bioAssembly);
// send some commands to Jmol
jmolPanel.evalString("select * ; color structure ; spacefill off; wireframe off; backbone off; cartoon on; select ligands ; spacefill 0.4; color cpk;");
System.out.println("done!");