MBTOpen:About
MBT Open
MBT Open is an open-source, Java-based protein visualization and analysis toolkit. The toolkit builds upon BioJava to provide classes for efficiently loading, managing and manipulating protein structure and sequence data. The MBT Open excels especially in providing a rich set of state-of-the-art 3d-accellerated graphical visualization components, as well as 2D visualization components which can be easily “plugged together” to produce applications having sophisticated graphical user interfaces. Yet, with all of the GUI components provided in the toolkit, the core data i/o and manipulation classes may be used to write completely non-graphical applications (say, for implementing pure analysis codes, or, for producing a non-graphical “back end” for web-based applications).
History
MBT Open originated from a mature library called the Molecular Biology Toolkit, developed under its own grant by John Moreland and others at the San Diego Supercomputer Center (SDSC). The project was then taken under the umbrella of the Protein Databank (PDB). Several web-deployed applications have been created and deployed to the main PDB website, as well as the IEDB website, and are being maintained both in their roles on the PDB website and as examples for developers of MBT Open projects. The PDB contributed the Molecular Biology Toolkit code to the BioJava effort in 2008 in order to make it more accessible to a larger audience of developers and users.
Goals
The MBT Open project seeks to contribute to the open-source Bioinformatics community in the following ways:
- For the Non-computational Biologist: Provide an easily configurable set of 3d protein visualization tools aimed at a variety of biological disciplines
- For the Bioinformatics tool developer: Provide a state-of-the-art, cross-platform library for structural biology visualization and analysis, with a robust plugin system to allow easy extension
- For the Bioinformatics researcher: Provide a forum and experimentation bed for 3d protein visualization techniques and other computational structural biology themes
Current Applications Based on MBT Open
- Available at the RCSB PDB:
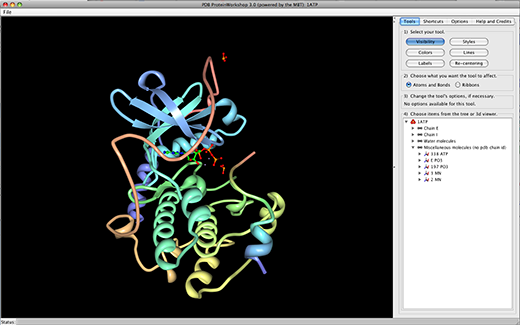
- Protein Workshop: a 3d protein viewer aimed at general viewing and publication-quality image generation
-
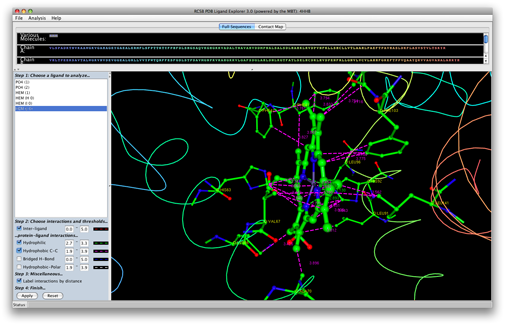
- Ligand Explorer: a 3d protein viewer aimed at visualization and analysis of ligands
-
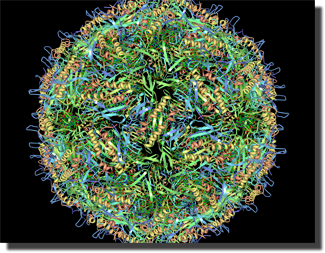
- Simple Viewer: a 3d protein viewer aimed at visualization of biological units and especially large repeated unit structures such as viral capsids
Source Code and Downloads
Source code and downloads will be available very soon.
How to Get Involved
Contributors needed! Instructions on how to get involved coming soon.
License
All MBT Open binaries and source code are released under the LGPL version 3 license.
Citations
When citing MBT, please reference J.L. Moreland, A.Gramada, O.V. Buzko, Qing Zhang and P.E. Bourne 2005 The Molecular Biology Toolkit (MBT): A Modular Platform for Developing Molecular Visualization Applications. BMC Bioinformatics, 6:21
–John Beaver 04:33, 12 November 2008 (UTC)